- 1 Department of Pathology and Molecular Diagnostics, Aichi Cancer Center, Nagoya, Japan
- 2 Department of Thoracic Surgery, Aichi Cancer Center Hospital, Nagoya, Japan
Lung cancer research has recently made significant progress in understanding the molecular pathogenesis of lung cancer and in developing treatments for it. Such achievements are directly utilized in clinical practice. Indeed, the echinoderm microtubule-associated protein-like 4–anaplastic lymphoma kinase (ALK) fusion gene was first described in non-small cell lung cancer in 2007, and a molecularly targeted drug against the fusion was approved in 2011. However, lung cancer with the ALK fusion constitutes only a small fraction of lung cancers; therefore, efficient patient selection is crucial for successful treatment using the ALK inhibitor. Currently, RT-PCR, fluorescent in situ hybridization (FISH), and immunohistochemistry are commonly used to detect the ALK fusion. Although FISH is currently the gold standard technique, there are no perfect methods for detecting these genetic alterations. In this article, we discuss the advantages and disadvantages of each method and the possible criteria for selecting patients who are more likely to have the ALK fusion. If we can successfully screen patients, then ALK inhibitor treatment will be the best example of personalized therapy in terms of selecting patients with an uncommon genotype from a larger group with the same tumor phenotype. In other words, the personalized therapy may offer a new challenge for current clinical oncology.
Introduction
Lung cancer research has recently made significant progress in understanding the molecular pathogenesis of lung cancer and in developing treatments for it. For example, epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (EGFR–TKIs) have been developed, and a subset of lung cancer patients have shown dramatic therapeutic responses to these treatments. Subsequently, this superior response was revealed to be associated with the EGFR mutation, which was identified in 2005 (Lynch et al., 2004; Paez et al., 2004). Currently, EGFR–TKIs are the first-line treatments for patients with advanced disease when the tumor is positive for the EGFR mutation. More recently, a similarly marked response to an anaplastic lymphoma kinase (ALK) inhibitor was demonstrated in patients with ALK fusion-bearing lung cancer (Kwak et al., 2010).
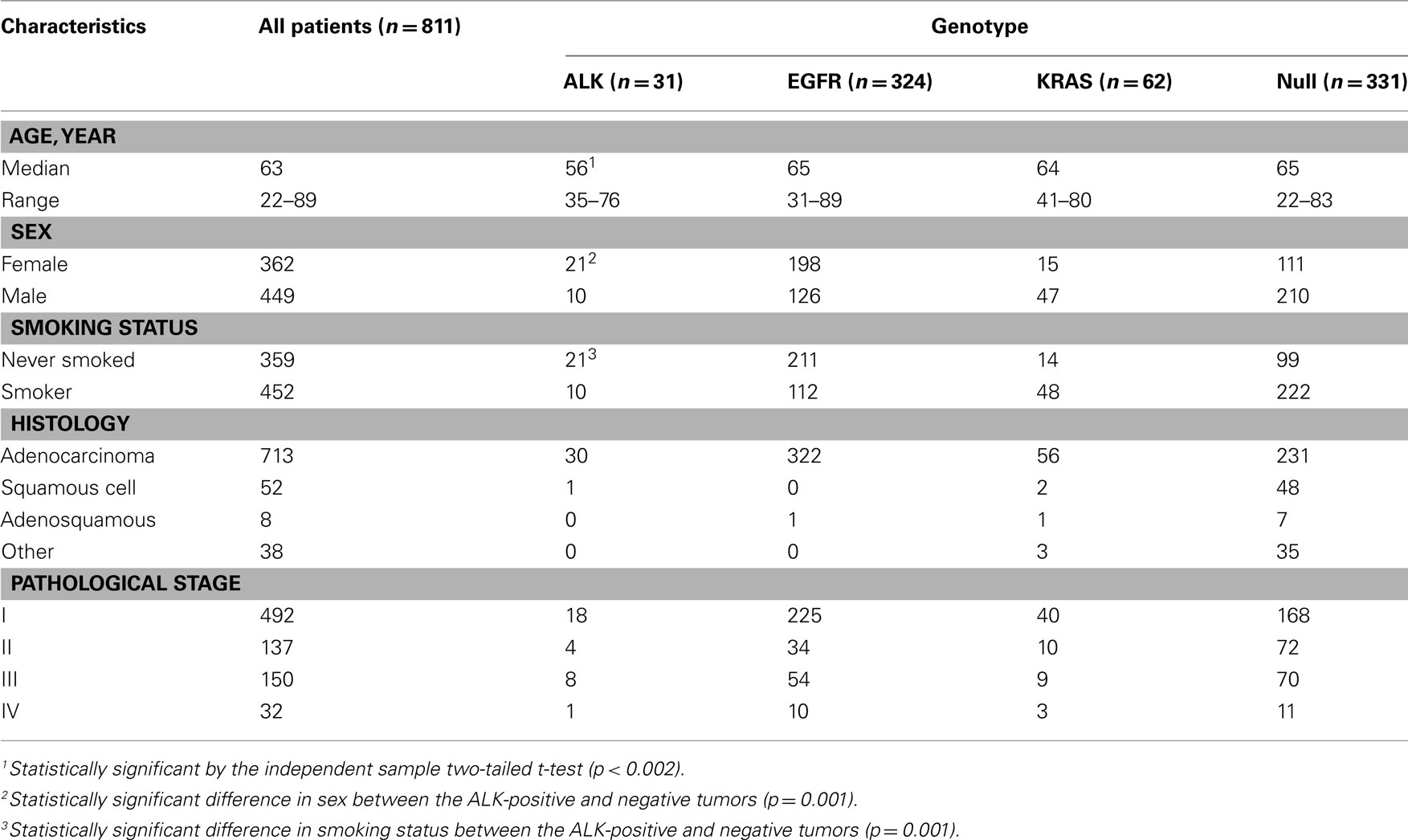
The echinoderm microtubule-associated protein-like 4 (EML4)–ALK fusion in non-small cell lung cancer (NSCLC) was first described in 2007 (Rikova et al., 2007; Soda et al., 2007). This fusion results from a small inversion within chromosome 2p that leads to the expression of a chimeric tyrosine kinase in which the N-terminal half of the EML4 is fused to the intracellular ALK kinase domain. The strong in vitro (Soda et al., 2007) and in vivo (Soda et al., 2008) oncogenic activity of the fusion protein has been demonstrated, and the inhibition of the fused molecule with ALK inhibitors leads to a marked deterioration of the tumor cells in vitro, supporting that ALK fusion is a driver mutation of lung cancer. A dramatic response to the ALK inhibitor similar to that of the EGFR–TKIs was expected and was observed. However, the prevalence of the fusion is low, ranging from 2 to 4% in the literature (Inamura et al., 2008; Koivunen et al., 2008; Perner et al., 2008; Shinmura et al., 2008; Takahashi et al., 2010) and in our consecutive series (Table 1). Even when the patients are selected for their predictive clinical characteristics, such as a lower median age and not having a history of smoking, it is difficult to identify the subsets of ALK-positive tumors. Therefore, efficient screening for the ALK fusion gene is a crucial issue in clinical practice.
Approximately one-third of lung cancer patients are diagnosed with localized disease that can be treated by surgical resection (Molina et al., 2006); however, nearly 30–35% will relapse after the initial surgery, even following a diagnosis at the earliest stage. For these patients, the surgically resected tissue may serve as an important sample for further molecular study because the recurrent tumor is often impossible to access without invasive procedures. In contrast, the other two-thirds of patients present with advanced, inoperable disease, and therefore a biopsy or a cytology specimen may be the only available tissue. Therefore, the methods used for detecting the ALK fusion should be applicable to these small tissue and cytology specimens. Currently, there are three methods for ALK testing: RT-PCR, fluorescent in situ hybridization (FISH) and immunohistochemistry. Because there is no perfect method, we need to know the applicable range of the samples and the sensitivity, specificity and limitations of the individual methods. Before explaining the details of these methods, we will consider whether the targeted patient population can be identified based on their clinicopathological features.
Concentration of the Patients with ALK-Positive Lung Cancer
Clinicopathological Features
Because ALK-positive lung cancer constitutes less than 5% of all lung cancers (Sasaki et al., 2010), it is critically important to select those patients who are more likely to have the ALK mutation. We initially attempted to select the patient population based on their clinicopathological features. As shown in Table 1, the patients with ALK-positive cancers were characteristically younger (a median age of 56 vs. 63), more frequently female, and more frequently non-smokers. In addition, the ALK fusion has a mutually exclusive relationship with the EGFR, KRAS, and HER2 mutations. Therefore, we attempted to select the ALK-positive lung cancer patients based on these clinicopathological features. The prevalence of the ALK translocation in the female patients younger than 63 without the EGFR, KRAS, HER2, or p53 mutations was 25% (Figure 1). However, four patients were not included in this subset (Mitsudomi et al., 2010). The selection of patients using clinicopathological features alone was able to increase the prevalence of the ALK mutation in our sample but was not able to identify all of the patients with it.
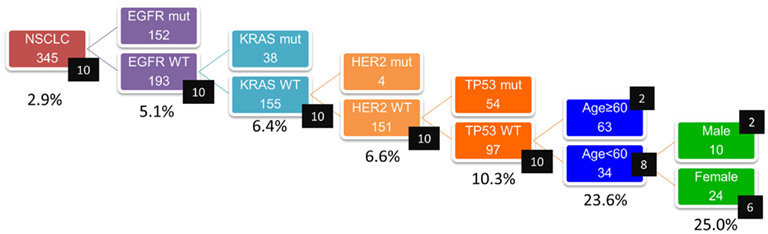
Figure 1. Concentration of EML4–ALK translocation using clinicopathologic features (n = 345). Numbers in black squares indicate number of patients with ALK translocation. Incidence of ALK translocation became 25% when patients were confined to those without EGFR, KRAS, HER2, p53 mutations and to female patients younger than 63. However, four patients were not included in this subset (Mitsudomi et al., 2010).
Morphological Features
An alternative method of selection uses the morphological features of the tumors. Table 2 contains several morphological features known to be characteristic of ALK-positive adenocarcinomas (Figure 2). These characteristics have been identified in studies comparing ALK-positive and ALK-negative lung cancers (Inamura et al., 2008; Rodig et al., 2009; Takeuchi et al., 2009; Yoshida et al., 2011), mostly using surgical specimens. Among these features, the presence of signet ring cells has the highest predictive value for ALK-positive lung cancer. According to Rodig et al. (2009) the ALK fusion was detected in 12 of the 26 tumors that they identified as having greater than 10% signet cells by area, although such tumors constituted only 8% of total lung adenocarcinomas. By contrast, the other morphological characteristics were more frequently seen in ALK-negative tumors. The solid-subtype was significantly more frequent in the ALK-positive cancers; however, the ALK-positive rate was 8% among the solid-subtype adenocarcinomas.
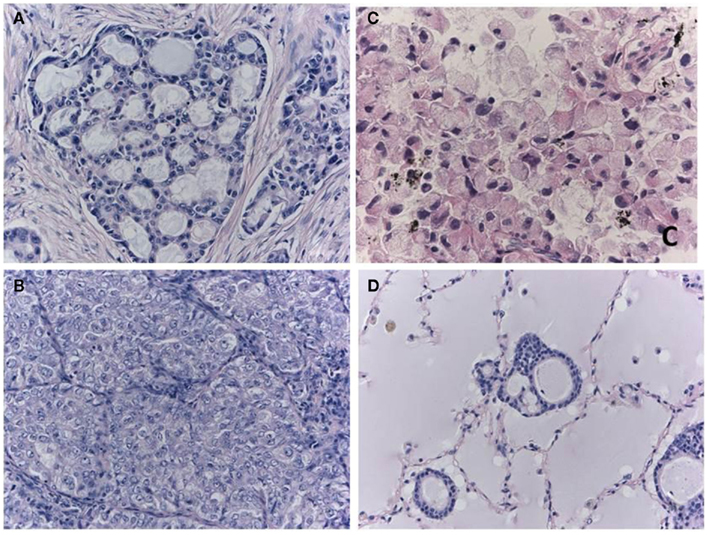
Figure 2. Morphological characteristics of ALK-positive lung cancer. The cribriform pattern (A) is composed of fused acini or glands. The solid-growth pattern consists of tumor cell nests without structural architecture (B). Solid adenocarcinoma cells with the ALK fusion often have vesicular nuclei with convoluted contours and deep grooves. This subtype may be misdiagnosed as squamous cell carcinoma or mucoepidermoid carcinoma in some cases. Signet ring cell carcinoma (C) is also characteristic of ALK-positive lung cancer. Rodig et al. (2009) found that 71% of the adenocarcinomas that consisted of more than 10% signet ring cells harbored the ALK fusion. One cellular characteristic of ALK-positive lung cancer is mucin production. In extreme cases, the alveolar space is filled with mucin (D), although the lumens of the neoplastic glands seldom open to the alveolar spaces. Although the majority of mucinous tumors are negative for TTF-1, the ALK-positive tumors (i.e., the signet ring cell carcinomas) are an exception.
In practice, candidate patients for ALK inhibitor treatments typically have advanced cancer, and their biopsy specimens and/or cytology are the major source of samples. Therefore, predicting these patients’ ALK fusion status based solely on morphological features is notably difficult.
Three Key Methods of Detecting ALK Fusion
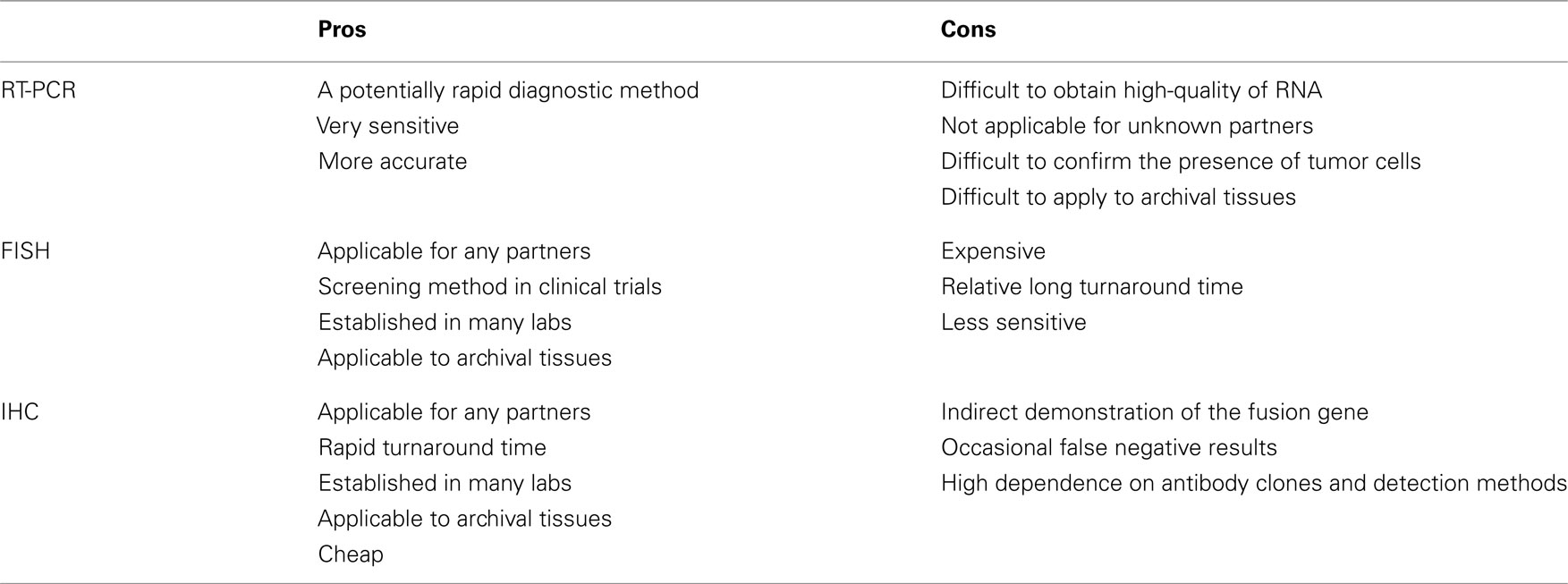
Currently, the methods of RT-PCR, FISH, and immunohistochemistry have been used to detect the ALK fusion gene. Because each method inevitably has both advantages and disadvantages (Table 3), we should be aware of their characteristics before applying them to clinical samples.
RT-PCR
RT-PCR is technically easy and rapid. The chromosomal inversion that characterizes the ALK fusion makes the sequence unique, and the PCR primer only hybridizes with the fusion chimeric transcript. This unique primer is responsible for the high sensitivity of this method. Soda et al. (2007) detected fusion mRNA in sputum that contained as few as 10 EML4–ALK-expressing BA/F3 cells. This method is used only for known fusion partners, however, and all 11 of the reported variants require skillful application of the technique because of the long PCR products. In addition, high-quality RNA is difficult to obtain in clinical practice. We found that 4 of the 361 tumors we screened had inconsistent RT-PCR and immunohistochemical results. Three of the tumors revealed both new and known variants of the EML4–ALK fusion transcript with 5′-RACE. Therefore, this method is difficult to implement in a routine clinical diagnostic laboratory. However, RT-PCR is one of the few methods that provide direct evidence of the chromosomal translocation. A demonstration of chimeric transcription is the best direct evidence of the translocation; if the results are negative, however, it is clinically difficult to determine whether the tumor is truly negative for the ALK fusion or whether it was not detected due to impaired RNA integrity or technical errors.
Fluorescent in situ Hybridization
Fluorescent in situ hybridization is currently the gold standard method used in clinical trials to detect the ALK fusion gene, and it was the first FDA-approved method for detecting the ALK fusion (Kwak et al., 2010). The FISH technique has been established in several labs to perform the HER2 test for breast cancer. FISH has the significant advantage of allowing archival material to be used. FISH also has disadvantages, however, including a relatively high cost and a long turnaround time. The evaluation of positive signals also requires considerable skill, especially when using biopsy samples. A break-apart FISH probe has been used to detect the ALK fusion (Figure 3), and the probes are designed for the telomeric and centromeric sides of the break points. Therefore, this design is applicable in detecting any kind of fusion partner, and any type of ALK gene rearrangement could theoretically be detected using this technique. However, it is known that some tumors with RT-PCR proven and ALK immunohistochemistry (IHC)-positive ALK fusions show non-split signals under the current criteria (Figure 4).
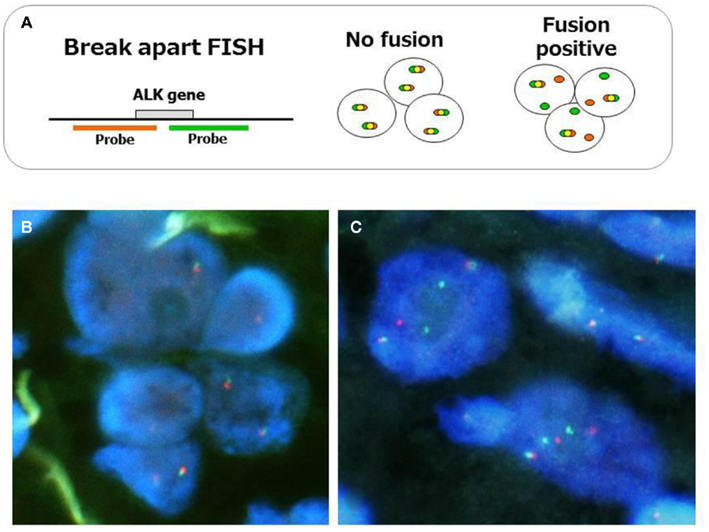
Figure 3. Fluorescent in situ hybridization method for ALK fusion detection. The probes, which are labeled with different fluorescences, are designed at telomeric and centromeric sides between break point (A). Representative FISH images of ALK-negative (B) and positive tumors (C) are displayed. When ALK is translocated, the signals are changed from merged yellow to single red and green. The current criteria of the gene rearrangement and positive ALK FISH are as follows. Cells are considered rearrangement positive when: (i) At least one set of orange and green signals are two or more signal diameters apart. (ii) There is a single orange signal without a corresponding green signal in addition to fused and/or broken apart signals. The tumor is considered positive ALK test when: (i) A sample is considered negative if <5 cells out of 50 (<5/50 or <10%) are positive. (ii) A sample is considered positive if >25 cells out of 50 (>25/50 or >50%) are positive. (iii) A sample is considered equivocal, if 5–25 cells (10–50%) are positive. If the sample is equivocal, a second reader should evaluate the slide.
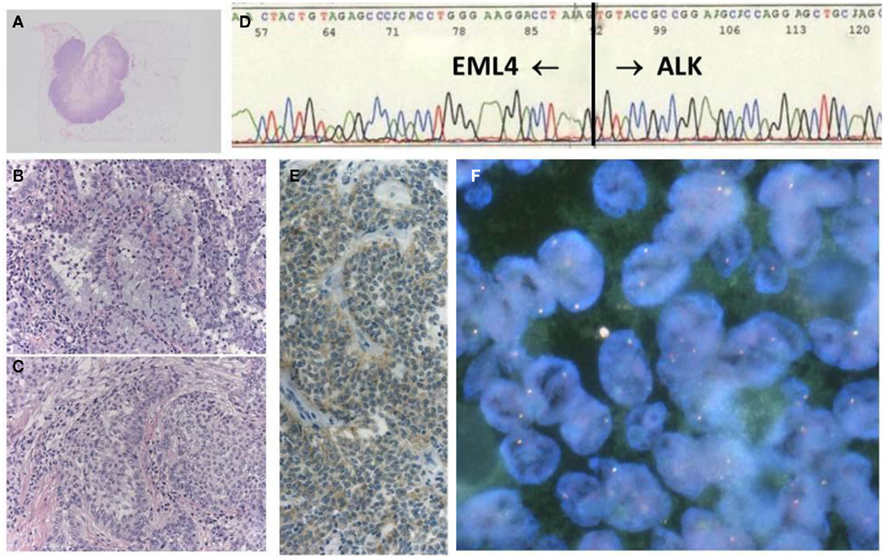
Figure 4. A case with EML–ALK-positive adenosquamous cell carcinoma, showing negative FISH. The tumor was a well-circumscribed nodule (A), in which both components of adenocarcinoma (B) and squamous cell carcinoma (C) were histologically identified. The chimeric transcript was detected with RT-PCR products (D), in addition to positive ALK-IHC (E). However, FISH results did not fulfill the positive criteria (F).
Immunohistochemistry
In contrast, immunohistochemical analysis is technically easy because it is integrated into routine pathological diagnosis. This assay has the advantages of rapidity, allowing the use of archival tissues, the ability to detect any partner genes, and simple comparisons of morphology. Although the immunohistochemical technique does not detect the ALK fusion gene itself, ALK is not detectable in any normal tissues other than the brain (Iwahara et al., 1997). Therefore, an ALK-positive reaction is associated with dysregulated expression of the gene, due to the altered promoter activity that is highly characteristic of an ALK inversion.
The early studies of ALK fusion immunohistochemistry reported that not all fusion-positive tumors yielded a positive immunohistochemistry result (Martelli et al., 2009). In contrast to the ALK fusion in anaplastic large cell lymphoma, the mRNA expression is lower in lung cancer; therefore, the false negative results appear to be caused by lower sensitivity. In general, the detection threshold is determined by the affinity of the primary antibody and the signal amplification system. In terms of the antibody affinity, Mino-Kenudson et al. (2010) compared two clones of the anti-ALK antibody and found large differences in their affinities for lung cancer cells. Nevertheless, it has been reported that highly sensitive immunohistochemistry is quite effective at detecting the ALK fusion gene. Takeuchi et al. (2009) compared the immunohistochemical results of combining three anti-ALK antibodies and two detection systems and found that all of the ALK-positive tumors could be detected by their highly sensitive immunohistochemical system, regardless of the differences between the clones. These two key factors, the antibody clones and the detection system, are important for detecting the ALK fusion by immunohistochemistry (Mitsudomi et al., 2011). Indeed, when we compared the results obtained using the ALK1 antibody with a conventional detection system to those obtained using the (high-affinity) 5A4 antibody with a highly sensitive detection system (Envision FLEX+ system), we found that 3 of 12 ALK-positive tumors went undetected by the former method (Table 4; Figure 5). 5A4 and D5F3 are known to be high-affinity antibody clones. When a highly sensitive detection system was used, either clone was able to detect all of the ALK-positive cancers, although D5F3 caused membranous staining in some ALK-negative tumors (Table 5).
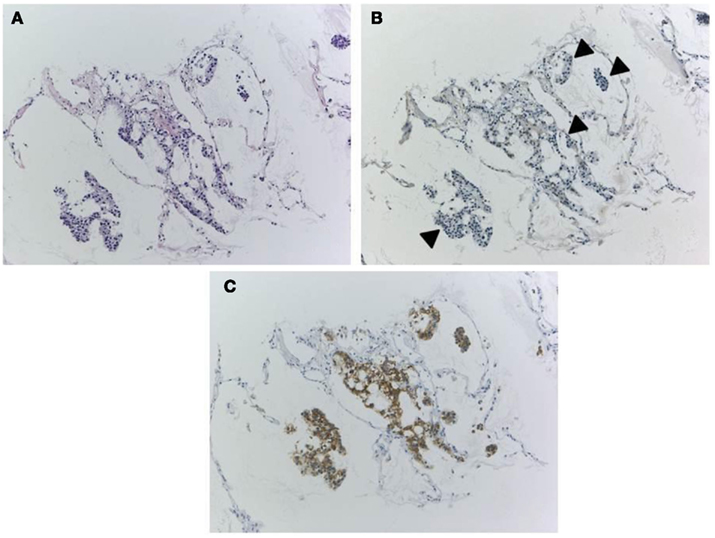
Figure 5. Discordant results according to antibody clones and detection system. A biopsy specimen only contains a small number of tumor cells [H&E staining, (A)]. When ALK1 antibody and conventional detection system was used, IHC showed negative results for tumor cells [(B), arrowheads]. However, proper clone (clone 5A4) and highly sensitive detection method (Envision FLEX+) made tumor cells show clearly positive reactions (C). Gene rearrangement and chimeric transcript of EML4–ALK fusion had been shown with FISH and RT-PCR in this specimen.

Table 4. Comparison between conventional (ALK1 and standard ABC methods) and optimized IHCs (5A4 and Flex+ system) using tissue microarray and biopsy specimens.
Because IHC does not directly demonstrate ALK fusion, there are certain pitfalls. Certain small cell lung cancers have positive reactions (Figure 6) but do not have the ALK translocation. This positive reaction did not appear to be associated with gene amplification, and the reason for the positive reaction was unknown. The ALK protein may be expressed in association with the neuroectodermal differentiation of small cell lung cancer, as it is expressed in the normal brain (Iwahara et al., 1997). Another pitfall is a certain rate of false negative reactions in signet ring cells, even when using a high-affinity antibody with a highly sensitive detection method. A large amount of cytoplasmic mucin can often push the cytoplasm into the rim, and the thinned cytoplasm weakly demonstrates a positive reaction in certain cases (Figure 7). Fortunately, pure signet ring cell carcinoma is extremely uncommon in lung cancer, and the other tumor components can show positive reactions, or a FISH analysis can be used instead.
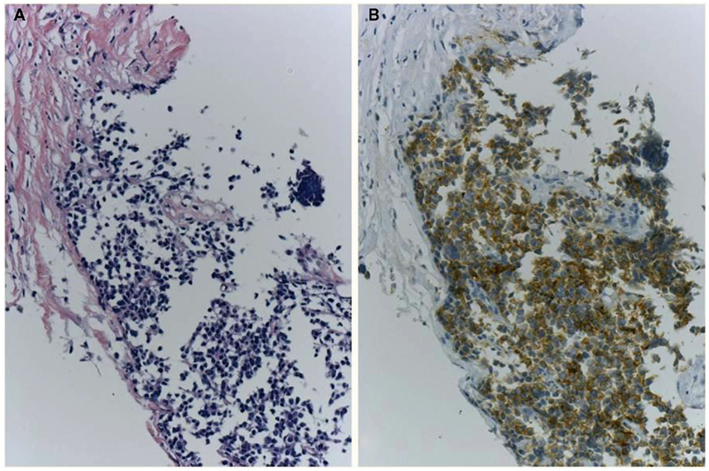
Figure 6. A case of ALK-IHC positive small cell lung cancer. A lung nodule was biopsied from the 83-year-old female smoker. H&E section (A) shows typical morphology of small cell lung cancer and CD56/NCAM and synaptophysin were positive. Although it is little reports of SCLC with ALK fusion, the tumor cells were positive for ALK immunohistochemistry using clone 5A4 and Envision FLEX+ (B). No gene rearrangement and chimeric transcript of EML4–ALK have been detected, respectively.
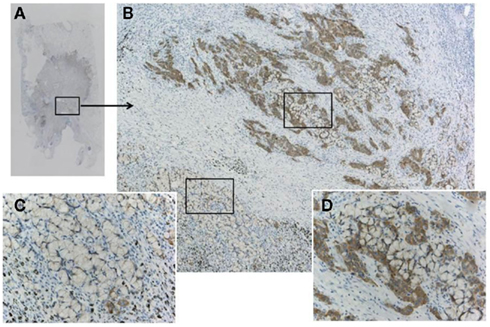
Figure 7. A pitfall of false negative immunohistochemistry in the mucinous component of adenocarcinoma, including signet ring cell carcinoma. Although the adenocarcinoma (A) was confirmed to harbor ALK fusion with both FISH and RT-PCR, immunohistochemistry using clone D5F3 and Envision FLEX+ showed various intensity of positive reactions (B). An upper part of the tumor cells (D) presented strong positivity, while the lower part showing signet ring cell carcinoma morphology appeared to be negative (C). Large amount of intracellular mucin pushes out cell cytoplasm outward to the rim. This false negative reaction does not indicate heterogeneity of ALK fusion, because split signals was demonstrated in both components.
ALK Testing Guidelines by the Japanese Lung Cancer Society
The ALK inhibitor crizotinib is expected to be introduced in Japan, and we face the practical application of the methods described above because this agent will be approved for patients with ALK fusion detected using any methods. This is in contrast with US policy, where crizotinib has been approved for ALK-positive patients only with an FDA-approved test. As noted, different precautions are necessary for individual methods as well as the appropriate handling of the tissues. Under the circumstances, the Japanese Lung Cancer Society issued a guideline for ALK testing (Figure 8). The guidance mentioned detailed the pros and cons of these methods, precautions for tissue handling, tissue application (biopsy, cytology, and surgical specimens) and comparisons of the three methods, including the different results of the various antibody clones against ALK for immunohistochemistry. Although validation of this algorithm has recently begun in a large prospective cohort, it is stressed that concurrent multiple methods should be used to select ALK-positive patients.
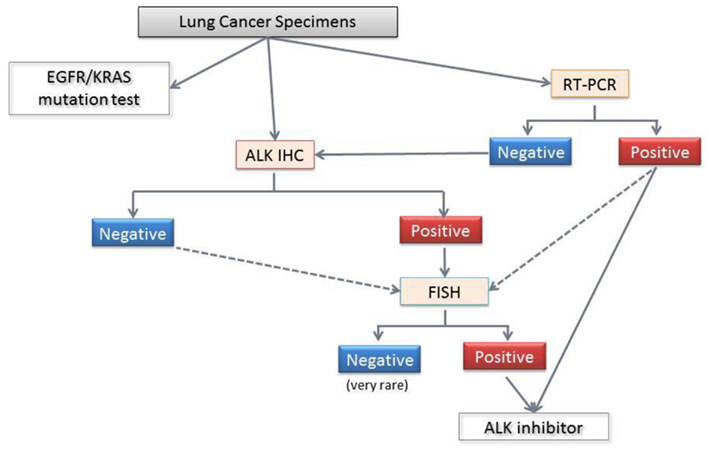
Figure 8. Diagnostic algorithm for ALK fusion from the Japanese Lung Cancer Society. The standard procedure for formalin-fixed paraffin embedded specimens involves screening with immunohistochemistry and confirmation with FISH. Even when the ALK immunohistochemistry is negative, FISH is recommended for the patients who are suspected of harboring the ALK fusion based on clinical considerations (age lower than 40 years, for example), and morphology (e.g., a mucinous cribriform pattern, signet ring cells, and TTF-1 positivity in mucinous adenocarcinomas). For pleural effusions and/or cytology samples, a positive RT-PCR can confirm the ALK fusion and directly facilitate crizotinib treatment. However, a negative RT-PCR does not have any clinical significance, and further examinations are required. Because each method has advantages and disadvantages, the use of more than one method to detect ALK fusion is recommended.
Conclusion
The lung cancer with ALK fusion constitutes only a few percentages in overall lung cancer, thus the target proportion is very limited in comparison with the other molecular targeted treatments. However, if we succeed to achieve successful screening of the patients, the treatment with ALK inhibitor will be the most representative personalized therapy in terms of selecting the patients from those harboring the same category of tumors according to the infrequent gene alteration. In other words, the achievement may be a new challenge for the current clinical oncology.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Noriko Shibata, Motoko Nimura, and Naoko Satoh for their excellent technical assistance with molecular, genetic, and immunohistochemical analyses.
References
Inamura, K., Takeuchi, K., Togashi, Y., Nomura, K., Ninomiya, H., Okui, M., Satoh, Y., Okumura, S., Nakagawa, K., Soda, M., Choi, Y. L., Niki, T., Mano, H., and Ishikawa, Y. (2008). EML4-ALK fusion is linked to histological characteristics in a subset of lung cancers. J. Thorac. Oncol. 3, 13–17.
Iwahara, T., Fujimoto, J., Wen, D., Cupples, R., Bucay, N., Arakawa, T., Mori, S., Ratzkin, B., and Yamamoto, T. (1997). Molecular characterization of ALK, a receptor tyrosine kinase expressed specifically in the nervous system. Oncogene 14, 439–449.
Koivunen, J. P., Mermel, C., Zejnullahu, K., Murphy, C., Lifshits, E., Holmes, A. J., Choi, H. G., Kim, J., Chiang, D., Thomas, R., Lee, J., Richards, W. G., Sugarbaker, D. J., Ducko, C., Lindeman, N., Marcoux, J. P., Engelman, J. A., Gray, N. S., Lee, C., Meyerson, M., and Janne, P. A. (2008). EML4-ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin. Cancer Res. 14, 4275–4283.
Kwak, E. L., Bang, Y. J., Camidge, D. R., Shaw, A. T., Solomon, B., Maki, R. G., Ou, S. H., Dezube, B. J., Janne, P. A., Costa, D. B., Varella-Garcia, M., Kim, W. H., Lynch, T. J., Fidias, P., Stubbs, H., Engelman, J. A., Sequist, L. V., Tan, W., Gandhi, L., Mino-Kenudson, M., Wei, G. C., Shreeve, S. M., Ratain, M. J., Settleman, J., Christensen, J. G., Haber, D. A., Wilner, K., Salgia, R., Shapiro, G. I., Clark, J. W., and Iafrate, A. J. (2010). Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N. Engl. J. Med. 363, 1693–1703.
Lynch, T. J., Bell, D. W., Sordella, R., Gurubhagavatula, S., Okimoto, R. A., Brannigan, B. W., Harris, P. L., Haserlat, S. M., Supko, J. G., Haluska, F. G., Louis, D. N., Christiani, D. C., Settleman, J., and Haber, D. A. (2004). Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 350, 2129–2139.
Martelli, M. P., Sozzi, G., Hernandez, L., Pettirossi, V., Navarro, A., Conte, D., Gasparini, P., Perrone, F., Modena, P., Pastorino, U., Carbone, A., Fabbri, A., Sidoni, A., Nakamura, S., Gambacorta, M., Fernandez, P. L., Ramirez, J., Chan, J. K., Grigioni, W. F., Campo, E., Pileri, S. A., and Falini, B. (2009). EML4-ALK rearrangement in non-small cell lung cancer and non-tumor lung tissues. Am. J. Pathol. 174, 661–670.
Mino-Kenudson, M., Chirieac, L. R., Law, K., Hornick, J. L., Lindeman, N., Mark, E. J., Cohen, D. W., Johnson, B. E., Janne, P. A., Iafrate, A. J., and Rodig, S. J. (2010). A novel, highly sensitive antibody allows for the routine detection of ALK-rearranged lung adenocarcinomas by standard immunohistochemistry. Clin. Cancer Res. 16, 1561–1571.
Mitsudomi, T., Suda, K., Tomizawa, K., and Yatabe, Y. (2010). Abstract #10598: Clinico-Pathologic Features of Lung Cancer with EML4-ALK Translocation. Available at: http://www.asco.org/ASCOv2/Meetings/Abstracts?&vmview = abst_detail_view&confID = 74&abstractID = 48829 [accessed].
Mitsudomi, T., Tomizawa, K., Horio, Y., Hida, T., and Yatabe, Y. (2011). Abstract #7534: Comparison of High Sensitive IHC, FISH, and RT-PCR Direct Sequencing for Detection of ALK Translocation in Lung Cancer. Available at: http://www.asco.org/ASCOv2/Meetings/Abstracts?&vmview = abst_detail_view&confID = 102&abstractID = 79101 [accessed].
Molina, J. R., Adjei, A. A., and Jett, J. R. (2006). Advances in chemotherapy of non-small cell lung cancer. Chest 130, 1211–1219.
Paez, J. G., Janne, P. A., Lee, J. C., Tracy, S., Greulich, H., Gabriel, S., Herman, P., Kaye, F. J., Lindeman, N., Boggon, T. J., Naoki, K., Sasaki, H., Fujii, Y., Eck, M. J., Sellers, W. R., Johnson, B. E., and Meyerson, M. (2004). EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 304, 1497–1500.
Perner, S., Wagner, P. L., Demichelis, F., Mehra, R., Lafargue, C. J., Moss, B. J., Arbogast, S., Soltermann, A., Weder, W., Giordano, T. J., Beer, D. G., Rickman, D. S., Chinnaiyan, A. M., Moch, H., and Rubin, M. A. (2008). EML4-ALK fusion lung cancer: a rare acquired event. Neoplasia 10, 298–302.
Rikova, K., Guo, A., Zeng, Q., Possemato, A., Yu, J., Haack, H., Nardone, J., Lee, K., Reeves, C., Li, Y., Hu, Y., Tan, Z., Stokes, M., Sullivan, L., Mitchell, J., Wetzel, R., Macneill, J., Ren, J. M., Yuan, J., Bakalarski, C. E., Villen, J., Kornhauser, J. M., Smith, B., Li, D., Zhou, X., Gygi, S. P., Gu, T. L., Polakiewicz, R. D., Rush, J., and Comb, M. J. (2007). Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 131, 1190–1203.
Rodig, S. J., Mino-Kenudson, M., Dacic, S., Yeap, B. Y., Shaw, A., Barletta, J. A., Stubbs, H., Law, K., Lindeman, N., Mark, E., Janne, P. A., Lynch, T., Johnson, B. E., Iafrate, A. J., and Chirieac, L. R. (2009). Unique clinicopathologic features characterize ALK-rearranged lung adenocarcinoma in the western population. Clin. Cancer Res. 15, 5216–5223.
Sasaki, T., Rodig, S. J., Chirieac, L. R., and Janne, P. A. (2010). The biology and treatment of EML4-ALK non-small cell lung cancer. Eur. J. Cancer 46, 1773–1780.
Shinmura, K., Kageyama, S., Tao, H., Bunai, T., Suzuki, M., Kamo, T., Takamochi, K., Suzuki, K., Tanahashi, M., Niwa, H., Ogawa, H., and Sugimura, H. (2008). EML4-ALK fusion transcripts, but no NPM-, TPM3-, CLTC-, ATIC-, or TFG-ALK fusion transcripts, in non-small cell lung carcinomas. Lung Cancer 61, 163–169.
Soda, M., Choi, Y. L., Enomoto, M., Takada, S., Yamashita, Y., Ishikawa, S., Fujiwara, S., Watanabe, H., Kurashina, K., Hatanaka, H., Bando, M., Ohno, S., Ishikawa, Y., Aburatani, H., Niki, T., Sohara, Y., Sugiyama, Y., and Mano, H. (2007). Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 448, 561–566.
Soda, M., Takada, S., Takeuchi, K., Choi, Y. L., Enomoto, M., Ueno, T., Haruta, H., Hamada, T., Yamashita, Y., Ishikawa, Y., Sugiyama, Y., and Mano, H. (2008). A mouse model for EML4-ALK-positive lung cancer. Proc. Natl. Acad. Sci. U.S.A. 105, 19893–19897.
Takahashi, T., Sonobe, M., Kobayashi, M., Yoshizawa, A., Menju, T., Nakayama, E., Mino, N., Iwakiri, S., Sato, K., Miyahara, R., Okubo, K., Manabe, T., and Date, H. (2010). Clinicopathologic features of non-small-cell lung cancer with EML4-ALK fusion gene. Ann. Surg. Oncol. 17, 889–897.
Takeuchi, K., Choi, Y. L., Togashi, Y., Soda, M., Hatano, S., Inamura, K., Takada, S., Ueno, T., Yamashita, Y., Satoh, Y., Okumura, S., Nakagawa, K., Ishikawa, Y., and Mano, H. (2009). KIF5B-ALK, a novel fusion oncokinase identified by an immunohistochemistry-based diagnostic system for ALK-positive lung cancer. Clin. Cancer Res. 15, 3143–3149.
Keywords: anaplastic large cell lymphoma kinase, non-small cell lung cancer, immunohistochemistry, RT-PCR, FISH, screening, molecular targeted drugs
Citation: Murakami Y, Mitsudomi T and Yatabe Y (2012) A screening method for the ALK fusion gene in NSCLC. Front. Oncol. 2:24. doi: 10.3389/fonc.2012.00024
Received: 05 December 2011; Accepted: 19 February 2012;
Published online: 16 March 2012.
Edited by:
Giuseppe Giaccone, National Institutes of Health, USACopyright: © 2012 Murakami, Mitsudomi and Yatabe. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Yasushi Yatabe, Department of Pathology and Molecular Diagnostics, Aichi Cancer Center, 1-1 Kanokoden, Chikusa-ku, Nagoya, Japan. e-mail: yyatabe@aichi-cc.jp
 Yoshiko Murakami1
Yoshiko Murakami1