- TB Immunology Laboratory, Department of Clinical Medicine, Institute of Molecular Medicine, Trinity College Dublin, Dublin, Ireland
miR-21 is one of the most highly expressed members of the small non-coding microRNA family in many mammalian cell types. Its expression is further enhanced in many diseased states including solid tumors, cardiac injury, and inflamed tissue. While the induction of miR-21 by inflammatory stimuli cells has been well documented in both hematopoietic cells of the immune system (particularly monocytes/macrophages but also dendritic and T-cells) and non-hematopoietic tumorigenic cells, the exact functional outcome of this elevated miR-21 is less obvious. Recent studies have confirmed a key role for miR-21 in the resolution of inflammation and in negatively regulating the pro-inflammatory response induced by many of the same stimuli that trigger miR-21 induction itself. In particular, miR-21 has emerged as a key mediator of the anti-inflammatory response in macrophages. This suggests that miR-21 inhibition in leukocytes will promote inflammation and may enhance current therapies for defective immune responses such as cancer, mycobacterial vaccines, or Th2-associated allergic inflammation. At the same time, miR-21 has been shown to promote inflammatory mediators in non-hematopoietic cells resulting in neoplastic transformation. This review will focus on functional studies of miR-21 during inflammation, which is complicated by the numerous molecular targets and processes that have emerged as miR-21 sensitive. It may be that the exact functional outcome of miR-21 is determined by multiple features including the cell type affected, the inducing signal, the transcriptomic profile of the cell, which ultimately affect the availability and ability to engage different target mRNAs and bring about its unique responses. Reviewing this data may illustrate that RNA-based oligonucleotide therapies for different diseases based upon miR-21 may have to target the unique and operative miRNA:mRNA interactions’ functionally active in disease.
Introduction
Micro-RNA-21 (miR-21) is an abundantly expressed microRNA in mammalian cells of multiple types (1–3). Its up-regulation is associated with many cancers, including those derived from both solid tissue (4, 5) and leukemic origin (6–9). The generation of a conditional miR-21 “knock-in” mouse confirmed that it functions as an oncogene with its overexpression resulting in malignant B-cell lymphoma (10). Functional studies performed in epithelia-, hepatocyte-, and glial cell-derived cell lines confirm that miR-21 regulates processes connected to cell growth, migration, and invasion (11–16), providing a mechanism for miR-21-mediated transformation of somatic cells. However, miR-21 is also expressed in hematopoietic cells of the immune system including B/T-cells, monocytes, macrophages, and dendritic cells (DCs). Activation of the immune system is strongly associated with tumor progression but also with surveying, responding to, and eliminating tumors as they arise. How increased miR-21 in these cell types facilitates tumor progression, as well as orchestrating the general immune response to pathogens and autoantigens in inflammatory disease, remains unclear. In this review, I will attempt to highlight some of the key findings on miR-21’s role in immunity and place this in the context of its dysregulation in disease including cancer and inflammation. I present a model of miR-21 as a key switch in immune circuits, controlling the balance between initial pro-inflammatory and later immuno-regulatory, anti-inflammatory responses, – dysregulation of which contributes to pathogenesis of inflammatory diseases including cancer and infection.
miR-21 Expression and Induction in Hematopoietic Cells
Immune Cell Maturation
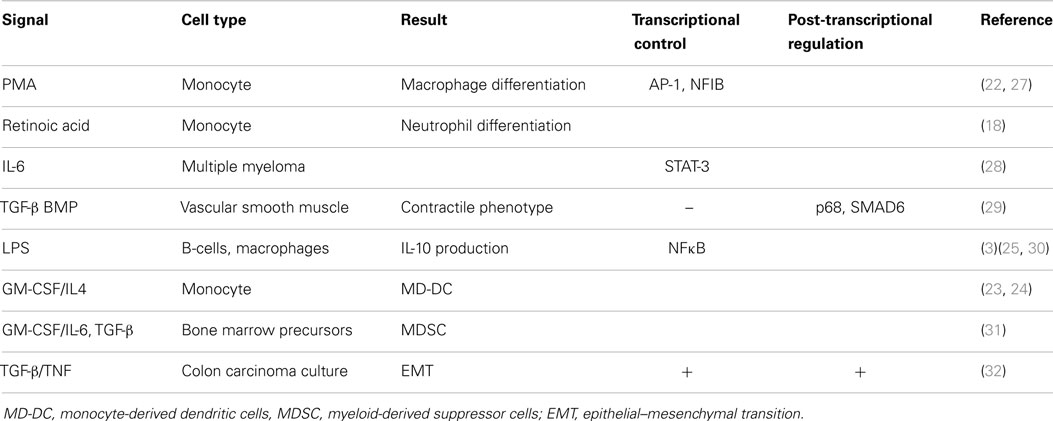
Initial efforts to profile miRNA expression during hematopoiesis revealed that while miR-21 is moderately expressed in hematopoietic progenitors, its expression increases significantly as various cell types mature to an “active” state, including bone marrow-derived mast-cells (17), neutrophils (18), and activated T-cells of various lineages (19, 20). High miR-21 levels are therefore a marker of immune cell activation, although whether or not this reflects a cause or consequence of activation remained to be determined. It was found that miR-21 expression is RNA polymerase II-dependent and derived from a primary transcript that is both capped and polyadenylated (21). Similar to regular protein-coding mRNAs, miR-21 expression is dynamically regulated by complex signaling pathways and can be enhanced by extracellular signals during immune cell development. The myeloid precursor cell type, the monocyte, which can be differentiated into various mature cells depending upon the extracellular signals received, shows increased expression of miR-21 during activation. This was first demonstrated by the study of Kashashima et al. (22), where TPA (also known as PMA) was used to differentiate monocytes toward macrophages. Since then, studies showing treatment of monocytes with all-trans retinoic acid to generate neutrophils (18), GM-CSF/IL-4 treatment to generate immature DCs (23, 24), and treatment with LPS (a TLR4 specific mimetic of bacterial endotoxin) to generate activated macrophages (25, 26), as well as LPS-mediated B-cell activation (3), all revealed significant up-regulation of miR-21. Table 1 lists these examples alongside many other immunologically relevant examples, but their detailed discussion is beyond the scope of this review. However, taken together, these data confirm that miR-21 serves as an important marker of immune cell activation in multiple contexts.
Turning the Circuit “on” – Induction of miR-21 by Inflammatory Stimuli
Like regular Pol-II-regulated protein-coding mRNAs, which are regulated by a diverse array of signal-specific transcription factors that bind unique sites at the promoter region, miR-21 exhibits diversity in the signals, transcription factors, and proposed binding sites that regulate its expression in diverse contexts. Unlike, regular Pol-II-regulated protein coding-genes and like all miRNAs, miR-21 is subject to an additional layer of post-transcriptional regulation before the mature 20 nt bioactive form is generated. This involves processing of both the precursor and mature duplex miRNA from the primary miRNA transcript (pri-miR-21), carried out by the nuclear enzyme Drosha and its cytosolic counterpart, Dicer. Many of the induction studies of miR-21 by extracellular signals including TPA/PMA, LPS, IL-6, and TGF-β/BMP have shown it to be a later event in their respective signaling pathways (25, 27–29). Expression analysis downstream of oncogene Ras-induced signaling, which drives miR-21 through AP-1, has shown that the appearance of mature bioactive miR-21 is delayed relative to the generation of pri-miR-21 (33). Although a dearth of studies have defined the role of various transcription factors in the induction of miR-21, including NFκB in LPS-induced miR-21 (25, 30), AP-1 in PMA-mediated up-regulation (27), and STAT-3 for IL-6-induced miR-21 (28), the complexity of the predicted promoter region of pri-miR-21 (27, 28) and the occurrence of alternative transcription start sites (34) suggest that the regulation of miR-21 transcription is not straight forward.
Several studies place miR-21 among the group of miRNAs whose post-transcriptional processing requires extra co-factors, notably the RNA helicase protein p68, which aids cleavage of the pri-miRNA transcript by Drosha (35). Although processing of mature miR-21 relative to other miRs may be differentially regulated by the inherent sequence differences of the miR-21 primary transcript recognized by Drosha/Dicer, a further layer of complexity is added when it is considered that the enzymes involved in miR-21 biogenesis are themselves regulated by extracellular signals, as shown in the study of TGF-β/BMP-mediated induction of mature miR-21. Here SMAD6, a key intracellular adapter protein activated by TGF-β signaling, bound to the primary miR-21 transcript and recruited p68 to promote Drosha-mediated cleavage during TGF-β signalling (29).
Interestingly, in a study of miR-21 induction in a model of colon carcinoma epithelial–mesenchymal transition (EMT), combined treatment with TGF-β and TNF induced pri-miR-21 and at a later stage, the appearance of the Drosha cleavage product, precursor-miR-21 stem-loop (32). This latter event required de novo protein synthesis and is indicative of an additional regulatory step to organize the temporal and cell-specific induction of miR-21. This important finding may be applicable to immune cells, which rapidly induce many cytokines and secreted factors, such as IL-6 or TNF, that have the potential to feed back and drive later events in the cell.
The story of miR-21 regulation grows even more complex when we consider the impact of other non-coding RNAs on its expression. A recent study illustrated that miR-21 is in fact regulated by a member of the long non-coding RNA family, GAS5 (36). Although lncRNAs can regulate genes at the transcriptional level, they also hold the potential to act as miRNA sponges – mopping up excessive mature 20 nt miRNAs and preventing them from engaging their mRNA targets. Therefore, GAS5 may in fact represent an important negative regulator of miR-21 activity, although this has yet to be examined in immune cells.
Expression in Diseased Tissue
Coincident with its induction in various immune cell types ex vivo, in vivo studies of diseased tissue often demonstrate increased expression of miR-21 relative to healthy control tissue. This has been shown in various models of allergic airway inflammation (26, 37), psoriasis and atopic eczema (38), osteoarthritis (39), and human atherosclerotic tissue (40), many of which are characterized by infiltration of immunocytes including macrophages, DCs, and T/B-cells. One can conclude from these studies and studies of miR-21 expression in cancerous tissue that increased miR-21 may act as a general biomarker of diseased tissue and in particular inflammation-associated diseases.
In a similar fashion, many studies of circulating miRNA profiles have implicated miR-21 as a secreted biomarker of disease due to its association with exosomes – small cell-derived vesicles whose cargo contains stable small RNAs including miRNA (41). Exosomes have been implicated as a mechanism of cell-to-cell communication and in this way act as classic immunomodulators. The fact that miR-21 is found in many exosomes including tumor-derived and immunocyte-derived (42, 43) supports a role for miR-21 as a key modulator of immune processes.
Few studies thus far have implicated the specific cell type responsible for the increased miR-21 expression in vivo. However, a recent study of miR-21 expression in gastric cancer found increased stromal, but not tumor cell, miR-21 to be strongly linked to clinical–pathological features of disease (44). In vivo analysis of mice challenged with Aspergillus fumigatus to model airway inflammation showed that miR-21 was induced in cells of the monocyte/macrophage lineage (26).
Inferring Function from Expression/Induction Studies
miR-21 is induced by many pro-inflammatory stimuli, both PAMPs and DAMPs, which trigger the inflammatory circuit and power up the cells of the immune system for action, illustrated in Figure 1A (immediate early response). However, the question remains as to what exact processes in this circuit this induced miR-21 regulates. The delayed induction of miR-21 in inflammatory reactions suggests that miR-21 may in fact negatively regulate the process of inflammation and be an important switch for the resolution of inflammation and maintenance of homeostasis, in essence counteracting the circuit, functioning as a trip-switch to turn off the often-damaging excessive pro-inflammatory response.
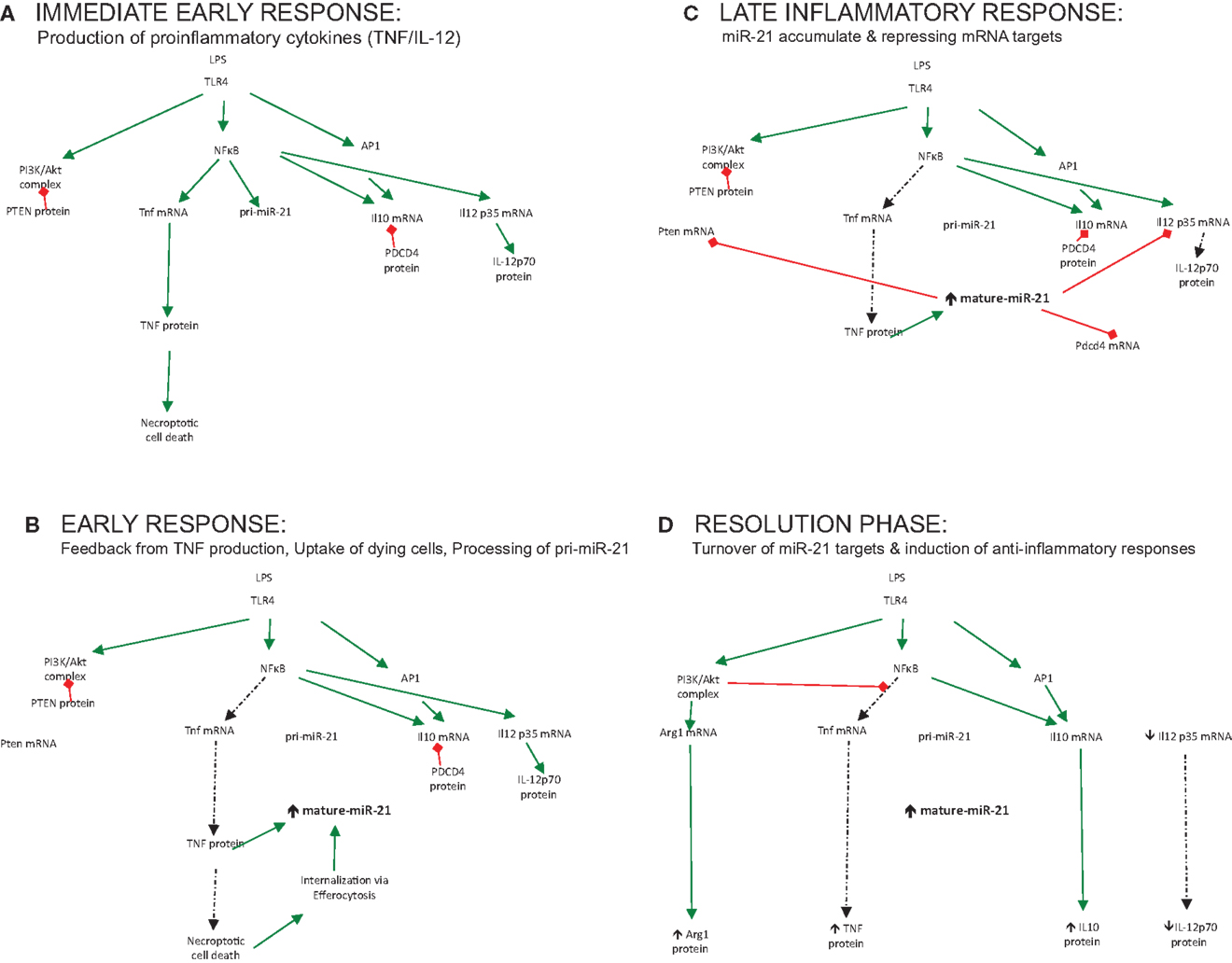
Figure 1. The role played by miR-21 regulating output of immune responses over time. (A) Immediate early response: production of proinflammatory cytokines (TNF/IL-12). (B) Early response: feedback from TNF production, uptake of dying cells, processing of pri-miR-21. (C) Late inflammatory response: miR-21 accumulate and repressing mRNA targets. (D) Resolution phase: turnover of miR-21 targets and induction of anti-inflammatory responses.
“Making the Switch” – Feedback of miR-21 as a Novel Regulator of Inflammatory Responses
The notion that miR-21 serves to limit inflammation and promote resolution should be supported by profiling studies of macrophage subsets; however, little induction of miR-21 is seen in alternatively-activated macrophages treated with IL-4 or IL-10 alone (45, 46). This supports the theory that an initial damage or danger signal needs to occur, which promotes an early pro-inflammatory stimulus such as NFκB or AP-1 to trigger miR-21. This ensures that miR-21 induction is appropriately activated to counteract damage triggered by infection. Recently, enhanced miR-21 expression was reported when LPS-treated macrophages were treated with apoptotic cells (47). This event fueled miR-21 expression to a greater extent, which was associated with decreased pro-inflammatory responses and the resolution of inflammation. Additionally, in an in vivo murine model of peritonitis, high level of miR-21 was reported, which was increased further following treatment with Resolvin D1, a lipid mediator that promotes resolution of inflammation (48). In this respect, damage or infection can be seen as the fuel that fires miR-21 expression and only when these have been appropriately sensed will miR-21 be appropriately up-regulated to counteract this, illustrated in Figure 1B (early response). This control mechanism ensures that miR-21 and its associated processes are not wastefully induced but “switched on” at appropriate times – when required to change the direction of the circuit and affect the balance of the inflammatory reaction to promote healing, resolution, and a return to homeostasis.
Ultimately, however, the function of a particular miRNA cannot be solely inferred from studies of its induction but must be deduced through studies of its activity also – namely, the specific mRNA targets it represses in any given context. For many miRNAs, bioinformatics analysis has aided the prediction and discovery of relevant mRNA targets. This computational-based approach has been less successful for miR-21, with many possible mRNA targets verified through various innovative techniques in cancer, inflammation, and other contexts, all of which will not be discussed here. Instead, I will limit the discussion to those identified targets and processes affected by miR-21, which tell us most about its role in immune responses (illustrated in Figure 1C – late inflammatory response) and discuss how these may promote the negative regulation of the inflammatory circuit, illustrated in Figure 1D (resolution phase).
PDCD4 and Cytokine Production
Our initial studies manipulating miR-21 during LPS signaling found that it had a unique effect on the levels of the anti-inflammatory cytokine IL-10, not observed for other cytokines. This was linked to the regulation of a proposed negative regulator of IL-10 production, PDCD4, loss of which was shown to protect from LPS lethality (25). Although the mechanism whereby PDCD4 regulates IL-10 and other cytokines remain an area of active investigation (49, 50), other groups have demonstrated that the miR-21/PDCD4 axis represents a key target for immunoregulation in multiple contexts, namely in protecting from type 1 diabetes (51), as a target for the endogenous danger ligand decorin-1 (52) and in regulating T-cell activation and polarization in SLE (53).
Recently the miR-21/PDCD4 axis was shown to play a key role in the process of efferocytosis (47) – the digestion and elimination of dead or dying cells by phagocytes, including macrophages, often associated with the induction of anti-inflammatory “clean-up” genes such as IL-10. Das et al. demonstrate that miR-21 levels are enhanced further in LPS-activated macrophages due to the uptake and internalization of apoptotic cells and, importantly, this process regulates IL-10 induction through PDCD4.
PTEN/PI3K Signaling
This study by Das et al. also clearly demonstrates a role for miR-21 in the regulation of TNF production, which separately from the miR-21/PDCD4 axis, is regulated by an additional miR-21 target gene, PTEN (47). A key intracellular kinase, PTEN is an important regulator of the PI3K/Akt pathway, which functions in many different cell types, each with unique functions and outcomes, but most strongly being pro-survival (54). It is not surprising then that elimination of PTEN, a negative regulator of PI3K activity, by dysregulated miR-21 would promote growth and survival in dividing somatic cells leading to malignant transformation (15, 55).
PTEN and PI3K signaling pathways have also been recently linked to macrophage phenotype and differentiation of functional subsets. Recently, studies of PTEN-deficient animals show more alternatively-activated macrophages in various models of polarization including Kuppfer cells, serving to protect from liver ischemia–reperfusion injury (56), as well as peritoneal macrophages marker expression (56, 57). The classical M2 marker Arg1, which is a key target for PI3K/Akt1 signaling, was found at much higher levels in these cells (57). Sahin et al. went on to demonstrate that this increased Arg1 expression resulted from activation of CEBPβ and STAT-3, as well as negative regulation of NFκB activity. Thus, miR-21 induction forms part of a key feedback circuit to limit excessive NFκB activity, turn off TNF production, and thereby transform the activated macrophage into a more reparative, “clean-up” cell, with key processes such as efferocytosis of dying cells, enhancing and promoting induction of this important immune modulator.
TNF Production
TNF has been associated with cell death and more recently high levels of TNF have been implicated in the death process observed in inflammatory macrophages labeled “necropoptosis” (58). Negative regulation of TNF levels by miR-21 therefore may not just help dampen down excessive inflammation but may also explain the effects of miR-21 on cell proliferation, migration, invasion, and transformation associated with excessive miR-21 levels and cancer. Strikingly, reciprocal regulation of miR-21 and TNF may in fact constitute an autoregulatory loop with evidence accumulating that TNF can promote miR-21 biogenesis (32) as well as the turnover of PDCD4 in macrophages (50). Moreover, the switch toward an anti-inflammatory, M2-like phenotype, and general immuno-regulatory environment, characterized by elevated IL-10 protein and increased Arg1 macrophage expression consistent with decreased TNF, which is associated with increased miR-21 (illustrated in Figure 1), may also account for poor immune responses against tumor-cells characteristic of tumor-associated macrophages (TAMs) found in cancer-induced stroma. Thus, stromal miR-21 induction (44) may constitute a pathogenic step in the biogenesis of cancer that leads to associated immunosuppression facilitating tumor growth and dissemination.
IL-12 and Antigen-Presenting Cells
The recent description of miR-21-deficient animals with profound defects in Th2 responses and skewing toward a Th1 response following administration of the OVA antigen (59) not only confirms the importance of miR-21 in directing T-cell polarization through its effects on innate antigen-presenting cells but also identifies T-cells and the adaptive immune response as a key target of miR-21 activity in immunity. Importantly, this study builds on earlier work (26) confirming an important target mRNA for miR-21; the IL-12p35 mRNA. IL-12, which acts as a strong inducer of Th1 responses and drives IFNγ production, is tightly controlled and in fact the p35 subunit is found at much lower levels in DCs and macrophages than its IL-12p70 partner, p40 (60). The finding that miR-21 can fine-tune its expression with big effects on subsequent immune responses in vivo, highlights the importance of this tiny 7 nt base-pair interaction. By directing the development of an appropriate T-cell response, this interaction again supports the notion that miR-21 controls the balance of pro- and anti-inflammatory responses. Accordingly, in diseases where miR-21 expression is dysregulated, this balance is altered with subsequent effects on innate but also adaptive immune cells, resulting in pathogenesis.
Impact of Dysregulated miR-21 on Immune Responses
Inflammatory Diseases
If miR-21 does indeed represent a key switch in the transition from a pro-inflammatory to an anti-inflammatory response, it stands to reason that at times, this key control point will become dysregulated with impact on the overall immune response, altering the control and balance of the whole circuit, which manifests as disease. As mentioned above, elevated miR-21 has been reported in many disease states. On the one hand, increased miR-21 expression is associated with conditions characterized by impaired immune responses including asthma (26), psoriasis (38), cancer (5), and importantly chronic bacterial or viral infections (61–66) (discussed below). Many of these conditions are associated with reprograming of pro-inflammatory M1 macrophages and/or Th1-cells and the appearance of regulatory immune cells including M2 macrophages, Th2, or regulatory T-cells. Therefore, miR-21 dysregulation by different triggers (DAMPs or PAMPs) may in fact promote disease pathogenesis by promoting an anti-inflammatory, immunosuppressive environment.
Conversely, increased miR-21 expression has also been reported in diseases fueled by chronic inflammation including colitis (67), atherosclerosis (40), type 2 diabetes (68), and SLE (53). In these cases, triggering a regulatory response through miR-21 would be beneficial, yet this is not manifested in the inflammatory environment of these diseased tissues. miR-21 up-regulation may simply be a marker of increased inflammation in these tissues, induced by the “fire” of the pro-inflammatory milieu. Curiously, ablation of miR-21 in some of these models, including colitis (67) and psoriasis (69), has actually been shown to offer protection from disease, indicating that miR-21 activity is promoting inflammation in these cases. In some cancer models, miR-21 expression itself is associated with inflammatory activation. It can promote NFκB activation in breast cancer cells (70) and TNF and IFNγ production in activated T-cells (71). Here, miR-21 is clearly acting to induce inflammation in transformed tumor-cells and activated T-cells rather than suppress inflammation in infected or activated macrophages. Differences in its function may relate to the different target mRNAs engaged in each cell type. Alternatively, miR-21 may augment general inflammation – both pro- and anti-inflammatory (50), with the reported effects on macrophage output observed after miR-21 modulation simply being reflective of other cues/signals in these cells at the same time.
Tumor Promoting Activity
Undoubtedly, miR-21 overexpression drives transformation of somatic cells and promotes tumorigenesis through effects on cellular growth, migration, and invasion (11–16). It is likely that the tumor microenvironment itself is also affected by miR-21 activity. As highlighted above, TAMs, which are re-programed from initial tumoricidal macrophages recruited to the site to immuno-permissive M2-like macrophages, are key cell types within the tumor microenvironment where miR-21 may be exerting pro-tumorigenic effects. Secretion of miR-21 from tumor-cell-derived exosomes or up-regulation of miR-21 in TAMs by tumor-derived pro-inflammatory products such as IL-6 or TNF, may participate in TAM reprograming and thereby facilitate growth, intravasation, and spread of tumor cells.
At the same time, as tumor-cells develop, intrinsic miR-21 may shape their responsiveness to therapy resulting in a more aggressive tumor phenotype. Alongside another immuno-responsive miRNA, miR-146, which functions as a negative regulator of TLR signaling pathways (72), miR-21 has been associated with chemoresistant ovarian epithelial cells (73). Importantly, these cells are characterized by low MyD88 expression and this key pro-inflammatory signaling protein has emerged as a key target for both miR-146 and miR-21. Low MyD88 expression in these aggressive cancer cells argues that TLR/IL-1 signaling may promote anti-cancer responses at this stage of disease and again dysregulation of miR-21 in the tumor acts to promote pathogenesis of disease progression.
Infection
As miR-21 regulates immune responses, it stands to reason that its induction may represent a target for subversion by invading pathogens in the ever-evolving arms race between the mammalian immune system and microbes. Indeed, many studies have characterized the rapid induction of miR-21 following infection of macrophages and other cells with microbes, including the pioneering work by Cameron et al. This study demonstrated that EBV induces miR-21 during latency, linking this miRNA with viral persistence (61). In a similar manner, infection of hepatocytes with either HBV or HCV was recently shown to induce miR-21 (62, 65), and in addition to promoting viral replication by enhancing growth and survival of the infected cell, miR-21 induction also modulates the host response in favor of the virus. Interestingly, signaling components of the TLR system (MyD88 and IRAK) have emerged from these studies as targets for miR-21 with the downstream consequence of decreased induction of anti-viral interferon-α during infection (62). Similarly, infection of renal cells with pseudorabies virus (PRV) induces miR-21, which targets mRNA for the important host chemokine CXCL10/IP-10 (63).
Studies of pathogen-induced miR-21 not only tell us more about the important immune-relevant target mRNAs for miR-21 but also about immune evasion strategies employed by the pathogen. For example, mycobacterial species, which persist and replicate in macrophages by successfully interfering with host responses, have been shown to induce miR-21. This subsequently targets multiple components of key pathways required for mycobacterial containment, including vitamin-D-dependent induction of anti-microbial peptides and the induction of pro-inflammatory cytokines including IL-1, TNF, IL-12, and IFNγ (64, 66). In particular, the finding that the avirulent mycobacterial strain BCG, used with mixed success to vaccinate against tuberculosis worldwide, induces miR-21 to escape immune responses (66), supports the notion that blocking miR-21 may in fact boost immunity and therefore temporal and specific inhibition of miR-21 may be an ideal candidate for vaccine development.
Rewiring the Circuit – miR-21 as an Attractive Target for Therapeutic Intervention?
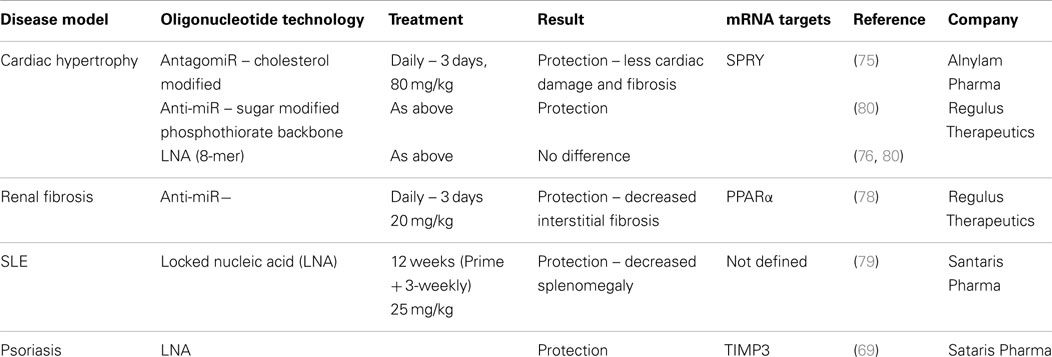
With interest in antisense technology increasing due to improved delivery techniques, specific targeting and more effective chemistries emerging, programs to target miR-21 in disease are being developed (74). Published studies have shown beneficial effects in various models although the exact mechanism contributing to this remains unclear. These studies are listed in Table 2, which highlights differences and commonalities in the methodologies and approaches used. Although effects of silencing miR-21 using antisense technology to counteract cardiac fibroblast remodeling in response to stress (75) were not reproducible in miR-21-deficient mice (76), there remains interest in blocking miR-21’s pro-fibrogenic activity particularly in response to ischemic–reperfusion injury (77). Early studies using anti-miR technology to block interstitial fibrosis demonstrated that protection from disease was generated through modulation of the key metabolic sensor, and miR-21 target, PPARα (78). However, it remains possible that miR-21 can exert some of its pro-fibrogenic activities through regulation of inflammatory signaling pathways such as IL-10 and TGF-β. Antisense to miR-21 has also been shown to reduce disease in two models of chronic inflammatory disease – psoriasis and SLE, with miR-21 inhibition in these cases apparently reducing inflammation, through effects on T- and B-cell activation and proliferation (in the SLE model) (79) and through negative regulation of MMP activity and TNF production (in the inflamed epidermis in the psoriasis model) (69).
Recent basic science studies attempting to understand miR-21’s complex biology better, are affecting targeting strategies and the development of miR-21 modulators for disease. Intriguingly, a study of miR-21 overexpression in hepatocytes observed differences in mRNA target engagement dependent upon the degree of overexpression, correlating with dysregulation of miR-21 in diseased tissue (81). This confirms that miR-21 behaves differently under various circumstances, including the level of miR-21 up-regulation itself and will affect strategies to target miR-21 in diseased tissue. With the widespread availability of advanced transcriptomic technologies, we may need to move toward a closer examination of elevated miR-21 and its impact upon the host cell transcriptome to get a clearer picture of the exact processes regulated by this particular miRNA.
As with any therapy designed to alter the balance of immune responses, the possibility of off-target effects or predisposition to other conditions, perhaps those characterized by chronic inflammation, exists. With greater understanding of miR-21 regulation and function, we may be able to tailor RNA-therapies and avoid off-target consequences. Furthermore, the notion of targeting specific miRNA:mRNA interactions via morpholino technology may avoid the deleterious effects of broad inhibition of miR-21 (82) and more specific delivery technologies such as β-glucan microparticles, which hone specifically to macrophages could be utilized to limit the effects of inhibition to a specific target cell-type (83). The transience and high turnover of RNA itself may help limit the effects of antisense treatment to the short-term. At the same time, caution needs to be erred when inhibiting an miRNA as ubiquitous and promiscuous as miR-21. Indeed, the area of miR-21 turnover and decay itself remains an unexplored area and its study may enhance our understanding of the role of miR-21 in immune responses, providing alternative means to antisense technology for limiting its expression in vivo.
Adaptive Immunity
Thus far, this review has concerned itself with miR-21 induction in cells of the innate immune system. However, as alluded to earlier, miR-21 is also found in both T and B-cells and its role in these cells is the subject of much investigation. Thus, employment of strategies to target miR-21 for modulation of immune responses requires anticipation of the effects on these cell types also.
Profiling studies of T-cells indicate that miR-21 is induced and acts as a marker of activated T-cells (19, 20), promoting survival and activation of these cells (84–86). In this way, miR-21 induction serves as a means to stratify naïve from activated T-cells, possibly assisting in the co-ordination of T-cell memory. Despite its expression across multiple T-cell subsets, intrinsic miR-21 can also affect T-cell polarization. Naïve T-cells transfected with miR-21 develop a more Th2/Treg phenotype (87) and this may be due to engaging different targets expressed in response to various other polarizing signals including BCL-6. T-cell miR-21 may also play an important role regulating tolerance to self, as demonstrated by studies showing exaggerated miR-21 induction in activated T-cells from PD1-deficient mice (88). These studies highlight the dual roles that T-cells play in regulating immune responses. While they must promote pro-inflammatory responses and eliminate infected cells, they must also orchestrate clearance of infection and promote resolution. T-cell miR-21 seems treads a fine line in balancing these processes and may become dysregulated during cases of autoimmunity.
Conclusion
Over the last 10 years, much effort has been placed in profiling the miR-nome of various cells under different conditions. From this, miR-21 has emerged as important miRNA both highly expressed and dynamically regulated in various cell types. Since then, identification of miR-21 function has been complicated not only by the possibility for many mRNA target interactions but also by its complex regulation in response to extracellular signals. The possibility has emerged that miR-21 can regulate numerous processes involved in correct cell function, survival, and proliferation, which if interrupted, can predispose to cellular transformation. However, it has also been linked to key processes involved in inflammation, detecting and responding to disturbances in homeostasis throughout the body, and orchestrating these responses appropriately. miR-21 therefore plays a dynamic role in inflammatory responses. Unlike other mediators, its presence is not solely characteristic of a pro-inflammatory or an immunosuppressive state, but is acting as a key signal mediating the balance and transition between both states. In essence, miR-21 induction can be seen as a “molecular rheostat” regulating the inflammatory switch. This makes it a novel and attractive target for therapeutic intervention and enhanced knowledge of its specific mRNA targets, as well as the signaling pathways and cellular processes regulated by miR-21, can only enhance its usefulness and attractiveness in this area.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The author would like to thank members of the TB Immunology Group and colleagues at the Trinity College, Dublin for reading and assistance in editing the manuscript, in particular Dr. Laura Elizabeth Gleeson. The author is supported by a Starting Investigators Research Grant (13/SIRG/2136) from Science Foundation Ireland.
References
1. Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science (2001) 294:853–8. doi:10.1126/science.1064921
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
2. Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol (2002) 12:735–9. doi:10.1016/S0960-9822(02)00809-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
3. Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell (2007) 129:1401–14. doi:10.1016/j.cell.2007.04.040
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
4. Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res (2005) 65:6029–33. doi:10.1158/0008-5472.CAN-05-0137
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
5. Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A (2006) 103:2257–61. doi:10.1073/pnas.0510565103
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
6. Fulci V, Chiaretti S, Goldoni M, Azzalin G, Carucci N, Tavolaro S, et al. Quantitative technologies establish a novel microRNA profile of chronic lymphocytic leukemia. Blood (2007) 109:4944–51. doi:10.1182/blood-2006-12-062398
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
7. Jongen-Lavrencic M, Sun SM, Dijkstra MK, Valk PJ, Lowenberg B. MicroRNA expression profiling in relation to the genetic heterogeneity of acute myeloid leukemia. Blood (2008) 111:5078–85. doi:10.1182/blood-2008-01-133355
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
8. Navarro A, Gaya A, Martinez A, Urbano-Ispizua A, Pons A, Balague O, et al. MicroRNA expression profiling in classic Hodgkin lymphoma. Blood (2008) 111:2825–32. doi:10.1182/blood-2007-06-096784
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
9. Pichiorri F, Suh SS, Ladetto M, Kuehl M, Palumbo T, Drandi D, et al. MicroRNAs regulate critical genes associated with multiple myeloma pathogenesis. Proc Natl Acad Sci U S A (2008) 105:12885–90. doi:10.1073/pnas.0806202105
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
10. Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature (2010) 467:86–90. doi:10.1038/nature09284
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
11. Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene (2007) 27(15):2128–36. doi:10.1038/sj.onc.1210856
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
12. Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem (2008) 283:1026–33. doi:10.1074/jbc.M707224200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
13. Gabriely G, Wurdinger T, Kesari S, Esau CC, Burchard J, Linsley PS, et al. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol (2008) 28:5369–80. doi:10.1128/MCB.00479-08
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
14. Lu Z, Liu M, Stribinskis V, Klinge CM, Ramos KS, Colburn NH, et al. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene (2008) 27:4373–9. doi:10.1038/onc.2008.72
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
15. Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology (2007) 133:647–58. doi:10.1053/j.gastro.2007.05.022
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
16. Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene (2007) 26:2799–803. doi:10.1038/sj.onc.1210083
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
17. Monticelli S, Ansel KM, Xiao C, Socci ND, Krichevsky AM, Thai TH, et al. MicroRNA profiling of the murine hematopoietic system. Genome Biol (2005) 6:R71. doi:10.1186/gb-2005-6-8-r71
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
18. Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature (2005) 435:834–8. doi:10.1038/nature03702
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
19. Cobb BS, Hertweck A, Smith J, O’Connor E, Graf D, Cook T, et al. A role for Dicer in immune regulation. J Exp Med (2006) 203:2519–27. doi:10.1084/jem.20061692
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
20. Wu H, Neilson JR, Kumar P, Manocha M, Shankar P, Sharp PA, et al. miRNA profiling of naive, effector and memory CD8 T cells. PLoS One (2007) 2:e1020. doi:10.1371/journal.pone.0001020
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
21. Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA (2004) 10:1957–66. doi:10.1261/rna.7135204
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
22. Kasashima K, Nakamura Y, Kozu T. Altered expression profiles of microRNAs during TPA-induced differentiation of HL-60 cells. Biochem Biophys Res Commun (2004) 322:403–10. doi:10.1016/j.bbrc.2004.07.130
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
23. Cekaite L, Clancy T, Sioud M. Increased miR-21 expression during human monocyte differentiation into DCs. Front Biosci (Elite Ed) (2010) 2:818–28. doi:10.2741/E143
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
24. Hashimi ST, Fulcher JA, Chang MH, Gov L, Wang S, Lee B. MicroRNA profiling identifies miR-34a and miR-21 and their target genes JAG1 and WNT1 in the coordinate regulation of dendritic cell differentiation. Blood (2009) 114:404–14. doi:10.1182/blood-2008-09-179150
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
25. Sheedy FJ, Palsson-McDermott E, Hennessy EJ, Martin C, O’Leary JJ, Ruan Q, et al. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat Immunol (2010) 11:141–7. doi:10.1038/ni.1828
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
26. Lu TX, Munitz A, Rothenberg ME. MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. J Immunol (2009) 182:4994–5002. doi:10.4049/jimmunol.0803560
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
27. Fujita S, Ito T, Mizutani T, Minoguchi S, Yamamichi N, Sakurai K, et al. miR-21 Gene expression triggered by AP-1 is sustained through a double-negative feedback mechanism. J Mol Biol (2008) 378:492–504. doi:10.1016/j.jmb.2008.03.015
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
28. Loffler D, Brocke-Heidrich K, Pfeifer G, Stocsits C, Hackermuller J, Kretzschmar AK, et al. Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood (2007) 110:1330–3. doi:10.1182/blood-2007-03-081133
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
29. Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature (2008) 454:56–61. doi:10.1038/nature07086
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
30. Zhou R, Hu G, Gong AY, Chen XM. Binding of NF-kappaB p65 subunit to the promoter elements is involved in LPS-induced transactivation of miRNA genes in human biliary epithelial cells. Nucleic Acids Res (2010) 38:3222–32. doi:10.1093/nar/gkq056
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
31. Li L, Zhang J, Diao W, Wang D, Wei Y, Zhang CY, et al. MicroRNA-155 and MicroRNA-21 promote the expansion of functional myeloid-derived suppressor cells. J Immunol (2014) 192:1034–43. doi:10.4049/jimmunol.1301309
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
32. Cottonham CL, Kaneko S, Xu L. miR-21 and miR-31 converge on TIAM1 to regulate migration and invasion of colon carcinoma cells. J Biol Chem (2010) 285:35293–302. doi:10.1074/jbc.M110.160069
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
33. Talotta F, Cimmino A, Matarazzo MR, Casalino L, De Vita G, D’Esposito M, et al. An autoregulatory loop mediated by miR-21 and PDCD4 controls the AP-1 activity in RAS transformation. Oncogene (2009) 28:73–84. doi:10.1038/onc.2008.370
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
34. Ribas J, Ni X, Castanares M, Liu MM, Esopi D, Yegnasubramanian S, et al. A novel source for miR-21 expression through the alternative polyadenylation of VMP1 gene transcripts. Nucleic Acids Res (2012) 40:6821–33. doi:10.1093/nar/gks308
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
35. Fukuda T, Yamagata K, Fujiyama S, Matsumoto T, Koshida I, Yoshimura K, et al. DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nat Cell Biol (2007) 9:604–11. doi:10.1038/ncb1577
36. Zhang Z, Zhu Z, Watabe K, Zhang X, Bai C, Xu M, et al. Negative regulation of lncRNA GAS5 by miR-21. Cell Death Differ (2013) 20:1558–68. doi:10.1038/cdd.2013.110
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
37. Moschos SA, Williams AE, Perry MM, Birrell MA, Belvisi MG, Lindsay MA. Expression profiling in vivo demonstrates rapid changes in lung microRNA levels following lipopolysaccharide-induced inflammation but not in the anti-inflammatory action of glucocorticoids. BMC Genomics (2007) 8:240. doi:10.1186/1471-2164-8-240
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
38. Sonkoly E, Wei T, Janson PC, Saaf A, Lundeberg L, Tengvall-Linder M, et al. MicroRNAs: novel regulators involved in the pathogenesis of psoriasis? PLoS One (2007) 2:e610. doi:10.1371/journal.pone.0000610
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
39. Zhang Y, Jia J, Yang S, Liu X, Ye S, Tian H. MicroRNA-21 controls the development of osteoarthritis by targeting GDF-5 in chondrocytes. Exp Mol Med (2014) 46:e79. doi:10.1038/emm.2013.152
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
40. Raitoharju E, Lyytikainen LP, Levula M, Oksala N, Mennander A, Tarkka M, et al. miR-21, miR-210, miR-34a, and miR-146a/b are up-regulated in human atherosclerotic plaques in the Tampere Vascular Study. Atherosclerosis (2011) 219:211–7. doi:10.1016/j.atherosclerosis.2011.07.020
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
41. Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol (2007) 9:654–9. doi:10.1038/ncb1596
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
42. Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, et al. MicroRNAs bind to toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci U S A (2012) 109:E2110–6. doi:10.1073/pnas.1209414109
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
43. Tian T, Zhu YL, Zhou YY, Liang GF, Wang YY, Hu FH, et al. Exosome uptake through clathrin-mediated endocytosis and macropinocytosis and mediating miR-21 delivery. J Biol Chem (2014) 289:22258–67. doi:10.1074/jbc.M114.588046
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
44. Uozaki H, Morita S, Kumagai A, Aso T, Soejima Y, Takahashi Y, et al. Stromal miR-21 is more important than miR-21 of tumour cells for the progression of gastric cancer. Histopathology (2014) 65(6):775–83. doi:10.1111/his.12491
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
45. Graff JW, Dickson AM, Clay G, McCaffrey AP, Wilson ME. Identifying functional microRNAs in macrophages with polarized phenotypes. J Biol Chem (2012) 287:21816–25. doi:10.1074/jbc.M111.327031
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
46. McCoy CE, Sheedy FJ, Qualls JE, Doyle SL, Quinn SR, Murray PJ, et al. IL-10 inhibits miR-155 induction by toll-like receptors. J Biol Chem (2010) 285:20492–8. doi:10.1074/jbc.M110.102111
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
47. Das A, Ganesh K, Khanna S, Sen CK, Roy S. Engulfment of apoptotic cells by macrophages: a role of microRNA-21 in the resolution of wound inflammation. J Immunol (2014) 192:1120–9. doi:10.4049/jimmunol.1300613
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
48. Recchiuti A, Krishnamoorthy S, Fredman G, Chiang N, Serhan CN. MicroRNAs in resolution of acute inflammation: identification of novel resolvin D1-miRNA circuits. FASEB J (2011) 25:544–60. doi:10.1096/fj.10-169599
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
49. van den Bosch MW, Palsson-McDermott E, Johnson DS, O’Neill LA. LPS induces the degradation of programmed cell death protein 4 (PDCD4) to release Twist2, activating c-Maf transcription to promote interleukin-10 production. J Biol Chem (2014) 289:22980–90. doi:10.1074/jbc.M114.573089
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
50. Yasuda M, Schmid T, Rubsamen D, Colburn NH, Irie K, Murakami A. Downregulation of programmed cell death 4 by inflammatory conditions contributes to the generation of the tumor promoting microenvironment. Mol Carcinog (2010) 49:837–48. doi:10.1002/mc.20660
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
51. Ruan Q, Wang T, Kameswaran V, Wei Q, Johnson DS, Matschinsky F, et al. The microRNA-21-PDCD4 axis prevents type 1 diabetes by blocking pancreatic beta cell death. Proc Natl Acad Sci U S A (2011) 108:12030–5. doi:10.1073/pnas.1101450108
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
52. Merline R, Moreth K, Beckmann J, Nastase MV, Zeng-Brouwers J, Tralhao JG, et al. Signaling by the matrix proteoglycan decorin controls inflammation and cancer through PDCD4 and microRNA-21. Sci Signal (2011) 4:ra75. doi:10.1126/scisignal.2001868
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
53. Stagakis E, Bertsias G, Verginis P, Nakou M, Hatziapostolou M, Kritikos H, et al. Identification of novel microRNA signatures linked to human lupus disease activity and pathogenesis: miR-21 regulates aberrant T cell responses through regulation of PDCD4 expression. Ann Rheum Dis (2011) 70:1496–506. doi:10.1136/ard.2010.139857
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
54. Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet (2006) 7:606–19. doi:10.1038/nrg1879
55. Ma X, Kumar M, Choudhury SN, Becker Buscaglia LE, Barker JR, Kanakamedala K, et al. Loss of the miR-21 allele elevates the expression of its target genes and reduces tumorigenesis. Proc Natl Acad Sci U S A (2011) 108:10144–9. doi:10.1073/pnas.1103735108
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
56. Yue S, Rao J, Zhu J, Busuttil RW, Kupiec-Weglinski JW, Lu L, et al. Myeloid PTEN deficiency protects livers from ischemia reperfusion injury by facilitating M2 macrophage differentiation. J Immunol (2014) 192:5343–53. doi:10.4049/jimmunol.1400280
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
57. Sahin E, Haubenwallner S, Kuttke M, Kollmann I, Halfmann A, Dohnal AB, et al. Macrophage PTEN regulates expression and secretion of arginase I modulating innate and adaptive immune responses. J Immunol (2014) 193:1717–27. doi:10.4049/jimmunol.1302167
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
58. Roca FJ, Ramakrishnan L. TNF dually mediates resistance and susceptibility to mycobacteria via mitochondrial reactive oxygen species. Cell (2013) 153:521–34. doi:10.1016/j.cell.2013.03.022
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
59. Lu TX, Hartner J, Lim EJ, Fabry V, Mingler MK, Cole ET, et al. MicroRNA-21 limits in vivo immune response-mediated activation of the IL-12/IFN-gamma pathway, Th1 polarization, and the severity of delayed-type hypersensitivity. J Immunol (2011) 187:3362–73. doi:10.4049/jimmunol.1101235
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
60. Kastelein RA, Hunter CA, Cua DJ. Discovery and biology of IL-23 and IL-27: related but functionally distinct regulators of inflammation. Annu Rev Immunol (2007) 25:221–42. doi:10.1146/annurev.immunol.22.012703.104758
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
61. Cameron JE, Fewell C, Yin Q, McBride J, Wang X, Lin Z, et al. Epstein-Barr virus growth/latency III program alters cellular microRNA expression. Virology (2008) 382:257–66. doi:10.1016/j.virol.2008.09.018
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
62. Chen Y, Chen J, Wang H, Shi J, Wu K, Liu S, et al. HCV-induced miR-21 contributes to evasion of host immune system by targeting MyD88 and IRAK1. PLoS Pathog (2013) 9:e1003248. doi:10.1371/journal.ppat.1003248
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
63. Huang J, Ma G, Fu L, Jia H, Zhu M, Li X, et al. Pseudorabies viral replication is inhibited by a novel target of miR-21. Virology (2014) 45(6–457):319–28. doi:10.1016/j.virol.2014.03.032
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
64. Liu PT, Wheelwright M, Teles R, Komisopoulou E, Edfeldt K, Ferguson B, et al. MicroRNA-21 targets the vitamin d-dependent antimicrobial pathway in leprosy. Nat Med (2012) 18:267–73. doi:10.1038/nm.2584
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
65. Qiu X, Dong S, Qiao F, Lu S, Song Y, Lao Y, et al. HBx-mediated miR-21 upregulation represses tumor-suppressor function of PDCD4 in hepatocellular carcinoma. Oncogene (2013) 32:3296–305. doi:10.1038/onc.2013.150
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
66. Wu Z, Lu H, Sheng J, Li L. Inductive microRNA-21 impairs anti-mycobacterial responses by targeting IL-12 and Bcl-2. FEBS Lett (2012) 586:2459–67. doi:10.1016/j.febslet.2012.06.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
67. Shi C, Liang Y, Yang J, Xia Y, Chen H, Han H, et al. MicroRNA-21 knockout improve the survival rate in DSS induced fatal colitis through protecting against inflammation and tissue injury. PLoS One (2013) 8:e66814. doi:10.1371/journal.pone.0066814
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
68. Zhong X, Chung AC, Chen HY, Dong Y, Meng XM, Li R, et al. miR-21 is a key therapeutic target for renal injury in a mouse model of type 2 diabetes. Diabetologia (2013) 56:663–74. doi:10.1007/s00125-012-2804-x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
69. Guinea-Viniegra J, Jimenez M, Schonthaler HB, Navarro R, Delgado Y, Concha-Garzon MJ, et al. Targeting miR-21 to treat psoriasis. Sci Transl Med (2014) 6:225re221. doi:10.1126/scitranslmed.3008089
70. Iliopoulos D, Jaeger SA, Hirsch HA, Bulyk ML, Struhl K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol Cell (2010) 39:493–506. doi:10.1016/j.molcel.2010.07.023
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
71. Ando Y, Yang GX, Kenny TP, Kawata K, Zhang W, Huang W, et al. Overexpression of microRNA-21 is associated with elevated pro-inflammatory cytokines in dominant-negative TGF-beta receptor type II mouse. J Autoimmun (2013) 41:111–9. doi:10.1016/j.jaut.2012.12.013
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
72. Boldin MP, Taganov KD, Rao DS, Yang L, Zhao JL, Kalwani M, et al. miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J Exp Med (2011) 208:1189–201. doi:10.1084/jem.20101823
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
73. d’Adhemar CJ, Spillane CD, Gallagher MF, Bates M, Costello KM, Barry-O’Crowley J, et al. The MyD88+ phenotype is an adverse prognostic factor in epithelial ovarian cancer. PLoS One (2014) 9:e100816. doi:10.1371/journal.pone.0100816
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
74. van Rooij E, Purcell AL, Levin AA. Developing microRNA therapeutics. Circ Res (2012) 110:496–507. doi:10.1161/CIRCRESAHA.111.247916
75. Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature (2008) 456:980–4. doi:10.1038/nature07511
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
76. Patrick DM, Montgomery RL, Qi X, Obad S, Kauppinen S, Hill JA, et al. Stress-dependent cardiac remodeling occurs in the absence of microRNA-21 in mice. J Clin Invest (2010) 120:3912–6. doi:10.1172/JCI43604
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
77. Xu X, Kriegel AJ, Jiao X, Liu H, Bai X, Olson J, et al. miR-21 in ischemia/reperfusion injury: a double-edged sword? Physiol Genomics (2014) 46(21):789–97. doi:10.1152/physiolgenomics.00020.2014
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
78. Chau BN, Xin C, Hartner J, Ren S, Castano AP, Linn G, et al. MicroRNA-21 promotes fibrosis of the kidney by silencing metabolic pathways. Sci Transl Med (2012) 4:121ra118. doi:10.1126/scitranslmed.3003205
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
79. Garchow BG, Bartulos Encinas O, Leung YT, Tsao PY, Eisenberg RA, Caricchio R, et al. Silencing of microRNA-21 in vivo ameliorates autoimmune splenomegaly in lupus mice. EMBO Mol Med (2011) 3:605–15. doi:10.1002/emmm.201100171
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
80. Thum T, Chau N, Bhat B, Gupta SK, Linsley PS, Bauersachs J, et al. Comparison of different miR-21 inhibitor chemistries in a cardiac disease model. J Clin Invest (2011) 121:461–2. doi:10.1172/JCI45938
81. Androsavich JR, Chau BN, Bhat B, Linsley PS, Walter NG. Disease-linked microRNA-21 exhibits drastically reduced mRNA binding and silencing activity in healthy mouse liver. RNA (2012) 18:1510–26. doi:10.1261/rna.033308.112
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
82. Choi WY, Giraldez AJ, Schier AF. Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science (2007) 318:271–4. doi:10.1126/science.1147535
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
83. Soto ER, Caras AC, Kut LC, Castle MK, Ostroff GR. Glucan particles for macrophage targeted delivery of nanoparticles. J Drug Deliv (2012) 2012:143524. doi:10.1155/2012/143524
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
84. Ruan Q, Wang P, Wang T, Qi J, Wei M, Wang S, et al. MicroRNA-21 regulates T-cell apoptosis by directly targeting the tumor suppressor gene Tipe2. Cell Death Dis (2014) 5:e1095. doi:10.1038/cddis.2014.47
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
85. Smigielska-Czepiel K, van den Berg A, Jellema P, Slezak-Prochazka I, Maat H, van den Bos H, et al. Dual role of miR-21 in CD4+ T-cells: activation-induced miR-21 supports survival of memory T-cells and regulates CCR7 expression in naive T-cells. PLoS One (2013) 8:e76217. doi:10.1371/journal.pone.0076217
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
86. Wang L, He L, Zhang R, Liu X, Ren Y, Liu Z, et al. Regulation of T lymphocyte activation by microRNA-21. Mol Immunol (2014) 59:163–71. doi:10.1016/j.molimm.2014.02.004
87. Sawant DV, Wu H, Kaplan MH, Dent AL. The Bcl6 target gene microRNA-21 promotes Th2 differentiation by a T cell intrinsic pathway. Mol Immunol (2013) 54:435–42. doi:10.1016/j.molimm.2013.01.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
88. Iliopoulos D, Kavousanaki M, Ioannou M, Boumpas D, Verginis P. The negative costimulatory molecule PD-1 modulates the balance between immunity and tolerance via miR-21. Eur J Immunol (2011) 41:1754–63. doi:10.1002/eji.201040646
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: miR-21, inflammation, cancer, PDCD4, macrophage
Citation: Sheedy FJ (2015) Turning 21: induction of miR-21 as a key switch in the inflammatory response. Front. Immunol. 6:19. doi: 10.3389/fimmu.2015.00019
Received: 02 October 2014; Accepted: 09 January 2015;
Published online: 29 January 2015.
Edited by:
Susan Carpenter, University of California San Francisco, USAReviewed by:
Paul Quax, Leiden University Medical Center, NetherlandsRyan Michael O’Connell, The University of Utah, USA
Gabrielle Fredman, Columbia University, USA
Copyright: © 2015 Sheedy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Frederick J. Sheedy, Trinity Center for Health Sciences, Institute of Molecular Medicine, St James’ Hospital, Room 0.50, Dublin 8, Ireland e-mail: fsheedy@tcd.ie
 Frederick J. Sheedy
Frederick J. Sheedy